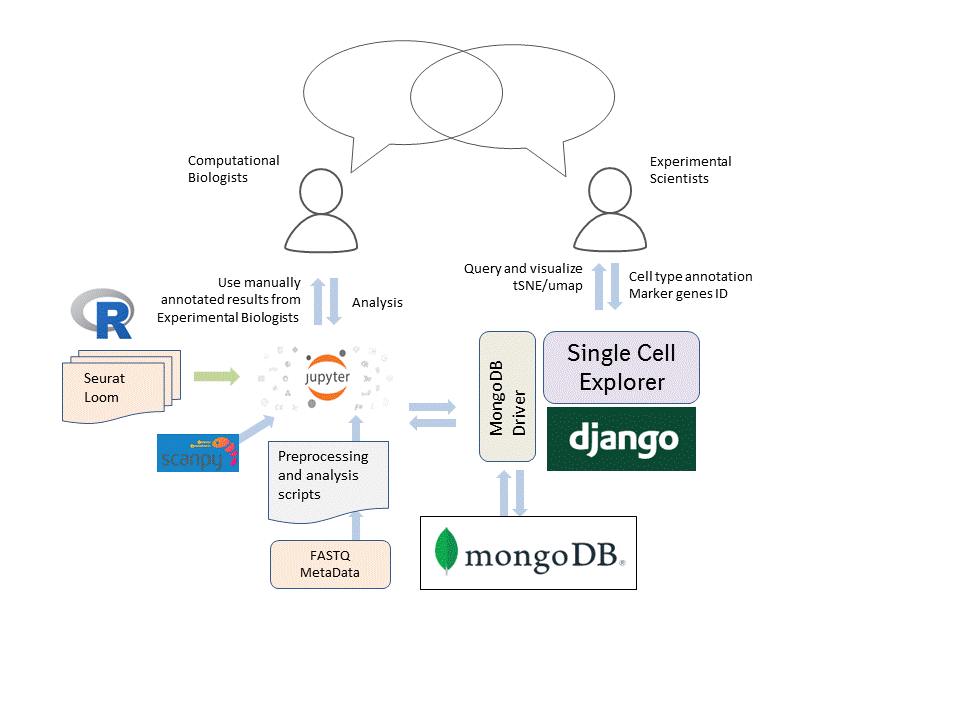
The mission of Single Cell Explorer is to facilitate crowdsourcing of data analysis, which will lead to novel discoveries in biology.
Our software allows experimental scientists to access information which normally requires coding skills - no coding necessary.
In doing so,
it streamlines the flow of information between computational biologists and experimental scientists.
With Single Cell Explorer, scientists can study and annotate interesting cell clusters based on t-SNE or UMAP,
using tools for freehand selection of cell clusters, checking cell marker expression,
and identifying unknown cell markers.
With Single Cell Explorer, multiple people can work on the same instance of a data set.
Any annotations or other work done by one person can be seen and edited by other collaborators,
so long as they have access to the instance.
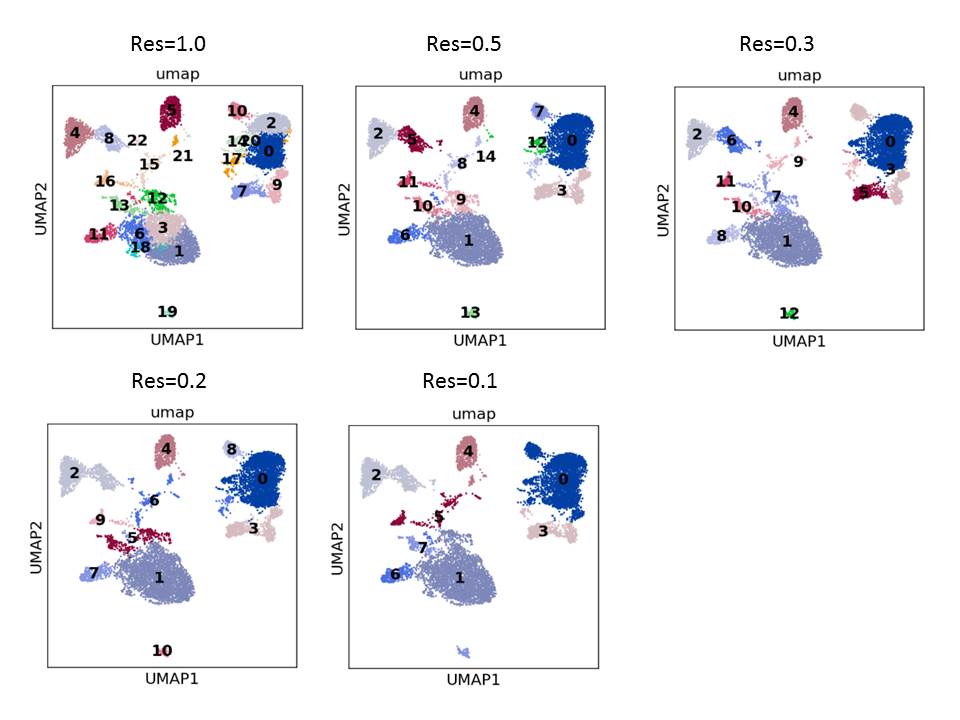
Choosing single cell cluster identification parameters can be ambiguous. The following plot shows various clustering results using different resolution parameters by the leiden algorhism (scanpy.api.tl.leiden function). It is difficult to determine which resolution to use. A resolution of 1, the default value, produces too many clusters in comparison with ground true.
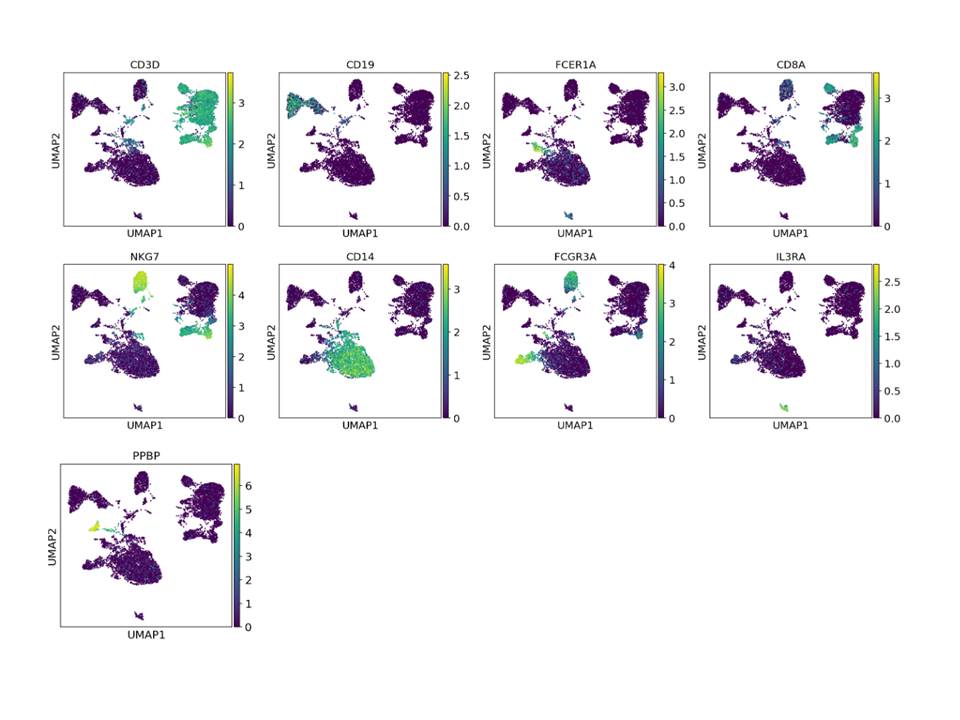
The ground truth can be reflected by the following UMAP, which was painted by known PBMC marker genes. This process can be done by biologists or domain experts (Immunologists) using Single Cell Explorer by leveraging prior knowledge. The result actually indicates a resolution of 0.2 might best reflect the current knowledge of PBMC cell type classification.